Previous lesson: Tidy Data and data wrangling
R programming basics: More data wrangling and piping
In this lesson, we will continue exploring ways to transform data frames by using conditional replacement. We will also use the ifelse() function to select between two alternatives. The module will conclude with the introduction of piping, a clean way to link a series of functions that we want to use to transform our data.
Learning objectives At the end of this lesson, the learner will be able to:
- use the
replace()function to change the value of a vector or data frame column. - use the
ifelse()function to produce varying outcomes depending on evaluating a condition. - modify tibbles by creating new columns or modifying previous ones.
- use pipes to simplify carrying out a linear sequence of data manipulations.
Total video time: 45 m 18 s
Links
Conditional replacement
The replace() function (8m07s)
The general format of the replace() function is
replace(source_vector, boolean_vector, replacement_value)
where source_vector and boolean_vector are the same length. The boolean_vector is usually generated by testing some condition. When the value for boolean_vector is TRUE for a particular item, the corresponding item in the source_vector is replaced by replacement_value. The returned value is a vector with the same length as the source_vector.
Problems with the replace() function (2m07s)
In this example, the replace() function is inadequate because we want two possible replacements.
Description of the ifelse() function (5m16s)
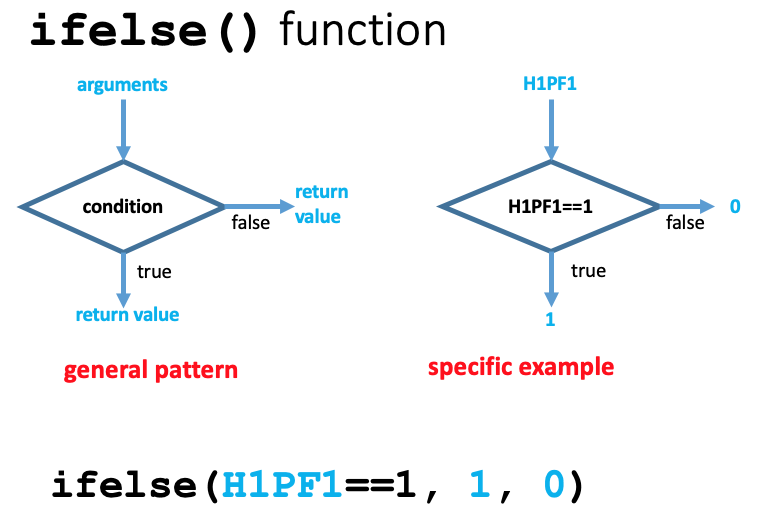
The general form of the ifelse() function is
ifelse(boolean_vector, value_if_true, value_if_false)
where boolean_vector is usually generated by testing some condition. The returned value is a vector with the same length as the boolean_vector. The diagram above shows the decision made for each value from a vector that is being tested for a condition. The returned value is a vector composed of a sequence made up from whichever of the two possible values corresponds to the evaluation of the condition for each position.
R scripting using the ifelse() function (3m25s)
In this example, we directly control the output using a vector of booleans
boolean_vector <- c(TRUE, FALSE, FALSE, TRUE, TRUE)
ifelse(boolean_vector, "yay!", "what?")
More typically, the boolean vector is generated by testing some condition involving another vector
ifelse(grades$participation=="pass", 100, 50)
In this case, we compare each character string in the participation column to the character string "pass".
Changing tibbles with ifelse() (7m06s)
In many functions, if a data frame is specified as the first argument, column names are assumed to correspond to that data frame.
mutate(grades, participation_numeric = ifelse(grades$participation=='pass', 100, 50)) # grades df specified in ifelse() function
mutate(grades, participation_numeric = ifelse(participation=='pass', 100, 50)) # grades df assumed in ifelse() function
When we carry out one of the replacement operations, we have choices about what do do with the output:
mutate(grades, participation_numeric = ifelse(participation=='pass', 100, 50)) # display in console
new_tibble <- mutate(grades, participation_numeric = ifelse(participation=='pass', 100, 50)) # assign to new tibble
grades <- mutate(grades, participation_numeric = ifelse(participation=='pass', 100, 50)) # assign back into the same tibble
grades <- mutate(grades, participation = ifelse(participation=='pass', 100, 50)) # replace the source column in the same tibble (change "in place")
Piping
Introducing data pipelines (6m30s)
A description of piping (6m14s)
In this example, the output of one function is stored temporarily in a data structure before being passed into the next function.
filename <- "https://gist.githubusercontent.com/baskaufs/ca8d32c1479de9e23cb93088ab8feef0/raw/1f94848c49f8b2e20e7bc93c890ac9caf5caa921/grades.csv"
grades <- read_csv(filename)
fixed_tests <- mutate(grades, tests = replace(tests, is.na(tests), 0))
fixed_participation <- mutate(fixed_tests, participation = ifelse(participation=='pass', 100, 50))
average_only <- transmute(fixed_participation, name, average = (tests + paper + participation)/3)
final_average <- filter(average_only, !is.na(average))
arrange(final_average, desc(average))
The output of the final function is sent to the console to be displayed to the user. Alternately, it could be assigned to a data frame by changing the last line to
summary <- arrange(final_average, desc(average))
Coding R with pipes (6m43s)
When data are piped into a function using the %>% pipe , the first argument specifying the input data object (often a data frame) doesn’t need to be specified.
read_csv("grades.csv") %>%
mutate(tests = replace(tests, is.na(tests), 0)) # no data frame name is specified
versus
grades <- read_csv(filename)
mutate(grades, tests = replace(tests, is.na(tests), 0)) # the "grades" data frame is specified
Somewhat surprisingly, the output of a pipe is assigned in the first line (not the last):
summary <- read_csv("grades.csv") %>%
mutate(tests = replace(tests, is.na(tests), 0)) %>%
mutate(participation = ifelse(participation=='pass', 100, 50)) %>%
transmute(name, average = (tests + paper + participation)/3) %>%
filter(!is.na(average)) %>%
arrange(desc(average))
The last step in a pipe can write to a file. In that case the output goes to the file rather than being displayed on the console.
Practice assignment
The practice assignment is here. You will need to load it into the editor pane of RStudio.
Problem 1 solution
Problem 2 solution
Problem 3 solution
Problem 4 solution
Problem 5 solution
Problem 6 solution
This is the last lesson in this module. Intermediate-level modules are under construction for Statistics with R and Data visualization using ggplot. Return to the CodeGraf landing page
Revised 2023-09-13

Questions? Contact us
License: CC BY 4.0.
Credit: "Vanderbilt Libraries Digital Lab - www.library.vanderbilt.edu"